


Isolation, Screening, and Characterization of L-Arginase Producing Soil Fungi in Saudia Arabia
Nourah Hassan Alzahrani
Department of Biology Sciences, Faculty of Science, University of Jeddah, Jeddah, KSA.
Abstract
Therapeutic enzymes have important roles in cancer treatment. Thus, our research target was to isolate fungi from Saudi soil; so examined their ability to produce L-arginase and identified them by 18S rRNA. From among the 20 fungal isolates recovered from the rhizosphere in the north and south of the city of Jeddah Province in Saudi Arabia, eight isolates produced high quantities of L-arginase. Highly produced fungal isolates were identified in the 18S rRNA sequence using DNA sequences of ITS1 and ITS4 and deposited in GenBank under joint numbers such as Aspergillus oryzaeXR_002735719.1, Penicillium oxalicum MH156644.1, Penicillium oxalicum MH171483.1, Aspergillus niger MF093522.1, Rhizopus oryzae AB250174.1, Aspergillus fumigates KJ746594.1, Fusarium equiseti KY945342.1, and Rhizopus oryzae AB250174. The highest producer of the enzyme was Fusarium equisetiKY945342.1 (2.492IU/mg & 1.952 IU/mg by Penicillium oxalicum MH156644.1).
Keywords: Fungi, L-arginase, 18S rRNA
L-Arginase (argininetransaminaseEC3.5.3.1) is a manganese-contained enzyme catalyzing L-arginine deamination to L-ornithine and urea and can be an anti-cancer agent and against tumors argininosuccinate synthase (Unissa et al., 2015). High rates of microbial generators and thermal reduction stabilities for enzyme could be the main biochemical barriers (El-Sayed et al., 2014). L-arginase is a therapeutic enzyme for orthostatic carcinogenic carcinomas, while low anti-cancer activities, low protein tolerances, and shorter serum half-life are its shortcomings (Hussain et al., 2017). L-arginase has strong anti-cancer activity against many tumors, by depleting arginine, which stops the cell cycle in stage G1. Arginine deiminases purify heat-loving Aspergillus fumigatus KJ434941 to its electrode homogeneity by 5.1 times, with a 50KD molecular subunit, a new thermostatic, less antigen and a strong candidate for clinical trials (Elsayed et al., 2015). L-arginase has counteracted a wide range of arginine auxotrophic cancers as hepatocellular carcinomas (Yoon et al., 2007; Kim et al., 2009; Glazer et al., 2011; Yau et al.,2010).
Many microbial strains have potentially produced enzyme and could have isolated and characterized. Enzymes can be produced by several microbial strains differed in physiological, biochemical, catalytic, and immunological characterization which leads to the continuous screening programsto isolate the novel microbial strains which might produce effective enzymes with little limitations in the usage sector (Unissa et al., 2015).
Different soil samples were collected from some plant rhizosphere from Jeddah province(Khulais &Bahra) from the surface and depth of 10 to 20 cm and were serially diluted. Aliquots of 0.1ml of the suspension were evenly spread on Modified Czapek Dox Agar. The plates were incubated at 28C for 72h.
Fungal isolates were appropriately screened for L-argininase production capacity by a rapid qualitative screening of the plates using (Czapek yeast extract agar medium) and phenol red (2g in 100ml ethanol) with last concentrations of 0.009% pre-poured plates as an indicator, yellow color zone around the colonies were reflected L-argininase enzyme and incubated in inverted position/ 28±2ºC for 48 to 96 hours. Production colonies were purified and sub-cultured from each plate for further studies.
Arginase activity was measured in terms of the hydrolysis rate from L-arginine to l-ornithine and urea by measuring the amount of urea released in the reaction. Urea was measured spectrophotometrically by the Archibald method (Unissa et al., 2015). Urea gave a dark pink color when heated in alcohol with α-isonitrosopropiophenone (0.2 ml glycine solution, 0.5 ml diluted enzyme & 0.1 ml manganese chloride). After incubation at 37ºC for 10 minutes, check the enzyme started by adding 0.1 ml of arginine was a mixture incubated again at 37ºC. After 30 minutes, the examination was stopped by adding 1 ml of a perchloric acid solution and mix.0.1 ml from 4% α-isonitrosopropiophenone added and tubes covered well and put in a boiling water bath. After 1 hour, the solution was removed and cooled at room temperature and was absorbed in540 nm. The urea produced from the prepared urea curve was estimated by changing the urea concentration between 0.1 to 1microliter\mol and a visual density graph was drawn against the urea concentration to extrapolate the arginase activity. The expression of one unit of L-arginase through the amount of enzyme which releases 1 µM of Lornithine under examination standard, based on the original Lornithine (El-Sayed, 2014).
Enzyme activity (µ/ml) = µmoles of urea released
time of enzyme action x volume of the enzyme (ml).
DNA isolated from fungal isolates using DNeasy® fungi Mini Kit
The DNA-amplified product was used for ITS1 and ITS4 use in PCR technology. The primer is designed according to Leio et al. (2016).
ITS1 (FORWARD) TCCGTAGGTGAACCTGCGG
ITS4 (REVERSE) TCCTCCGCTTATTGATATGC
The obtained sequence results were subject to a BLAST search program to identify similarities with genes in the gene bank.
Based on serial dilution 22 fungal isolates were obtained.
The primary screening of fungal isolates show their abilities for L-arginase activities carried out on modified MCD with arginine as nitrogen source using phenol red as the pH indicator.
Isolates show a yellow zone around colonies indicated decreasing in pH. The pink color turned to yellow in medium supplemented with L-Gln which means producing L-arginase, the yellow zone has appeared in plate number AAB1,AAD2,AAD4,AAB1,AAD3, AFA1, AFA3(Fig.1).
The selected fungal isolates were assayed quantitatively for L-arginase production for 7 days of incubation. It was noticed that all the fungal isolates produced L-arginase after three days of incubation, these amounts increased by increasing the incubation period, but the maximum amount of the enzyme was accumulated after five days of incubation by all of the fungal isolates. Results in Fig. (2) show the highest producer AFA3 2.492IU/mg, followed by isolate AAB4 1.952 IU/mg.
|
L-arginase Activity of Fungal Isolates on MCD Plates |
||
|
|
|
|
|
|
|
|
|
|
|
|
Figure 1: Qualitative Assay for L-arginase Activity of Fungal Isolates on MCD Plates
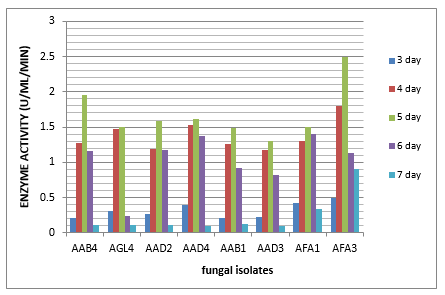
Figure 2: Quantitative Assay of L-Arginase Activity Produced by the Selected Fungal Isolates by Nessler Reaction
The fungal isolates were subjected to molecular analysis and identification using DNA sequencing of ITS1 and ITS4 primers (Cheng et al., 2016). First, the 750 bp amplicons were separated on 1% agarose gels to prove their accuracy and specificity. Then, the fragments with the molecular size representing the ITS4 and ITS1primers were purified for sequencing (Table 1).
|
Fungal Isolate |
Name and Accession of No. of the most related Strain in NCBI GenBank |
Fungal Isolates |
Identity(%) |
Coverage (%) |
|
AAB1 |
XR_002735719.1 |
Aspergillus oryzae |
100% |
100% |
|
AAB4 |
MH156644.1 |
Penicillium oxalicum |
100% |
100% |
|
AAD2 |
MH171483.1 |
Penicillium oxalicum |
100% |
100% |
|
AAD3 |
MF093522.1 |
Aspergillus niger |
100% |
100% |
|
AAD4 |
AB250174.1 |
Rhizopus oryzae |
100% |
100% |
|
AFA1 |
KJ746594.1 |
Aspergillus fumigates |
100% |
100% |
|
AFA3 |
KY945342.1 |
Fusarium equiseti |
100% |
100% |
|
AGL4 |
AB250174.1 |
Rhizopus oryzae |
100% |
100% |
Table 1: DNA Sequencing of 18s rRNA Using Universal Primer for Fungal Isolates
Screening enzymatic activity by various methods such as measuring the color change region or the filtration region on agar with the completion of the appropriate substrate was used (Balagurunathan et al., 2010b, Nagaraju and Ram, 2019). Phenol red shows the basic pH change which is red in alkaline condition turning to yellow under acidic condition (Theantana et al., 2007). Isolates show yellow color surrounding colonies which indicate pH reduction; these results agree with the observation of Gulati et al. (1997) and Jyothi (2011).
Nadaf et al. (2019) have screened newer strains able to fabricate new L-arginase that had bug yield and few adverse effects. Twenty isolates confirmed a red zone around the colonies, indicating that they produce L-arginase. There are only a few investigations for resources for the manufacturing of this enzyme. There are only a few investigations about sources of producing this enzyme. Pink color surrounded colonies are due to the liberation of L-arginase (Zhang et al., 2013).
According to Nocker et al. (2004) amplification, ITS1, and ITS4 sequences of fungi can produce 750bp amplicon. The DNA sequences were analyzed using Blast alignment tools of GenBank and showed that the eight isolates identified as Aspergillus oryzaeXR_002735719.1 were, Penicillium oxalicum MH156644.1, Penicillium oxalicum MH171483.1, Aspergillus niger MF093522.1, Rhizopus oryzae AB250174.1, Aspergillus fumigates KJ746594.1, Fusarium equiseti KY945342.1, Rhizopus oryzae AB250174.1 with the similarity percentages of 100%. Using ITS1 and ITS4 as semi-nested PCR performed positively on samples by 18S rRNA PCR (Ferrer et al., 2001). Our results are consistent with those of Anand et al., (2001) who found PCR examination effectiveness compared to traditional methods. The PCR-based test is a fast, sensitive, and useful method for detecting fungal isolates.
Enzyme activities were determined in cultures both using Nessler's reagent (Dhevagi and Poorani, 2006). All the fungal isolates produced L-arginase after three days of incubation, these amounts increased by increasing the incubation periods, but the maximum amount of the enzyme was accumulated after five days of incubation by all of the fungal isolates; the highest producer of the enzyme was Fusarium equisetiKY945342.1 AFA3 (2.492IU/mg and1.952 IU/mg by Penicillium oxalicum MH156644.1 (AAB4). Previous studies have reported the highest L-arginase activity of Bacillus thuringiensis SK 20.001, and Rummelii baciluspycnus SK31.001 that were8.4 U/ml 21 at 24 h and 40.2 U/ml 27 at 16 h respectively with an optimal temperature of 80℃ (Nadaf et al., 2019). Very few reports on prokaryotic L-rginase have been characterized (Zolfaghar et al., 2019) such as Bacillus brevis (Kanda et al., 1997), B. anthraci (Viator et al., 2008), and Helicobacter pylori (Zhang et al., 2011). Studies of fungi enzyme such as Neurospora crassa (Borkovich and Weiss 1987), Aspergillus nidulans (Borsuk et al., 1999), and Penicillium chrysogenum (El-Sayed et al., 2014) have been reported. Micro-organisms can be easily cultivated and manipulated, hence they are considered as good sources for enzymes production (Kumar and Sobha 2012).
The current study refers to the result of testing the production of the enzyme L-arginase by twenty fungal isolates obtained from different soil samples collected from the rhizosphere of a number of plants in Jeddah, Saudi Arabia. The quantitative and qualitative results showed the high ability of the fungal isolates to produce the enzyme which is one of the most important candidate enzymes for use in treating types of cancer. Fungal isolates were molecularly defined by 18S rRNA It can be concluded from this study that the rooting fungicide activity test has enzyme activity and the extract is expected to be used as therapeutic, industrial and anti-tumor agents.
REFERENCES
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.