


DNA Barcoding And Molecular Taxonomy Of Gracilaria Fergusonii J.Ag. Using rbcL Gene
J. John Peter Paul1*, C. Iniya Udhaya2
1 Assistant Professor of Botany and Director, Centre for Advanced Research in Plant Sciences (CARPS), Department of Botany, St. Xavier’s College (Autonomous), Palayamkottai - 627 002, Tamil Nadu, India.
2 Assistant Professor, Department of Botany, Sri Kaliswari College (Autonomous), Sivakasi - 626 123, Tamil Nadu, India.
ABSTRACT
DNA barcoding can be utilized as a contemporary tool for species identification and molecular phylogeny. It can decrease many important issues of morphological taxonomy, a small amount of tissue is also required for the identification of species, and the samples can be observed at all stages of growth. DNA barcoding can be utilized in the identification of invasive and endangered species along with conservation biology. In the case of red marine macroalga, DNA barcoding can be helpful for finding high yielding agar as well as for obscure species identification. In the present investigation, the red algae Gracilaria fergusonii J.Ag. was provided from Hare Island in Thoothukudi district, Tamil Nadu, India. DNA was extracted by Cetyl-Trimethyl-Ammonium-Bromide (CTAB) procedure. DNA segments coding for rbcL gene were amplified from total genomic DNA with oligonucleotide primers. The sequencing reaction was carried out in a PCR thermal cycler, utilizing the BigDye Terminator v3.1 cycle sequencing kit. From the present data, the chloroplast marker, rbcL gene is more effective for the DNA barcoding and molecular taxonomy of red marine macroalga.
Keywords: DNA barcoding, Molecular taxonomy, rbcL gene, Algae, Gracilaria fergusonii
Marine life is attractive and is considered to have a great perspective on the inherent value as well as for the progress of novel drugs. Marine flora and fauna are explained to have a wide spectrum of bioactive secondary metabolites and novel compounds that are structurally and biologically active (Aswal et al. 1984). In this era of globalization, human activities are resulting in climate change, pollution, coastal degradation, and the introduction of alien and invasive species. Among the various algae, the Rhodophyta or red algae are very significant, ecologically as well as commercially. Therefore, to investigate the distribution of algae, the identification of organisms is very important which is generally according to external characteristics. This is the traditional way of identification which is a little complex owing to its three major limitations (Hebert et al. 2003; Pires and Marinoni, 2010). Firstly, the morphological characters are not complete (May, 1988) as some groups (flowering plants and vertebrates) are better assessed than others (algae, nematodes). Secondly, the identification characters require well-trained taxonomists who become not always available for regular identifications. Thirdly, the organism to be identified may be small or at a growth stage, where the trained taxonomists may face difficulty in characterization and identification. Moreover, the morphological-based identification techniques are time-consuming and sometimes these data may do not provide classification up to the species level (Rindi et al. 2008; Packer et al. 2009).
During the last few decades, small segments of DNA, called DNA barcodes are used (El-Hamshary et al., 2018; El-Hamshary et al., 2019; Torabizadeh and Hashemi, 2019). DNA barcoding is a technique, which gives a fast identification of species without involving the morphological characteristics. It can utilize a fairly DNA fragment as an identifier, to describe or determine a species. In plants, the mitochondrial genome evolves much more slowly than in animals. The mitochondrial gene COXI region is not proper for the identification of plants (Rubinoff 2006). Thus, the plant DNA barcoding studies were primarily limited to the chloroplast genome to understand and compare the variation of the gene coding (matK, rbcL and rpoC1) and non-coding sequences (ITS and psbA-trnH), which were summarized by Chen et al. (2010). Lahaye et al. (2008) reported that the chloroplast genome of plants revealed a high degree of variation and discrimination potential. Among the coding sequence rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit) gene is sequenced for phylogenetic analysis of plants (Schuettpelz et al. 2006). Hence, our investigation aimed to determine the molecular identification, genetic relationships, and development of DNA markers of Gracilaria fergusonii J.Ag. utilizing rbcL sequencing.
Gracilaria fergusonii J.Ag. was collected from Hare Island (Thoothukudi district), the southeast coast of Tamil Nadu, India. Marine algae were collected on the low and subtidal regions up to 1m depth by handpicking. The collected algae were cleaned thoroughly with marine water in the field to discard other epiphytes and sediment soil particles. The plant samples were brought to the laboratory in wet conditions and washed again by distilled water to remove the salt on the surface of the plants. The marine macroalgae were identified by referring to the keys given by Umamaheswara Rao (1970 and 1987).
Genomic DNA was extracted from the young thalli of the chosen Gracilaria species based on the modified CTAB method utilized by Doyle and Doyle (1987 and 1990) with minor modifications.
Before the PCR analysis, the isolated DNA extractions were diluted 1:100 with sterile double distilled water. DNA coding fragments of rbcL gene were amplified from total genomic DNA with rbcL primers (Table 1) and PCR buffer, 0.2mM of each dNTPs, 1µl DNA, DNA polymerase enzyme, BSA and DMSO, Betaine, forward and reverse primers (Table 2). PCR parameters included an initial denaturing temperature of 98°C for thirty seconds and 40 repeated cycles of 98°C for 5 seconds, 58°C for 10 seconds, and 72°C for 15 seconds. The final incubation at 72°C for 60 seconds was applied to ensure complete polymerization of DNA strands (Table 3).
Table 1: Primers used in PCR amplification of rbcl gene
|
Target |
Primer Name |
Direction |
Sequence (5’ à 3’) |
|
rbcL |
Dt-rbcL-F56 |
Forward |
AGTGACCGTTACGAATCTGG |
|
Dt-rbcL-R1010 |
Reverse |
AGGATCACCTTCTAATTTACC |
Table 2: PCR Amplification conditions
|
Constituents |
Quantity |
|
MgCl2 |
1.5mM final conc. |
|
dNTPs 0.2Mm (each) |
4µl |
|
DNA |
1µl |
|
Phire Hotstart II DNA polymerase enzyme |
0.2µl |
|
BSA |
0.1mg/ml |
|
DMSO |
3% |
|
forward and reverse primers |
5pM |
|
Betaine |
0.5M |
Table 3: PCR Temperature profile
|
Profile |
Specification |
|
Initial denaturation |
98°C for thirty seconds |
|
Denaturation |
98°C for 5 seconds |
|
Annealing |
58°C for 10 seconds |
|
Extension |
72°C for 15 seconds |
|
Final extension |
72°C for 60 seconds |
|
Number of cycles |
40 cycles |
The sequencing reaction was carried out using PCR thermal cycler (GeneAmp PCR System 9700) in the BigDye Terminator v3.1 sequencing kit following the manufacture's protocol (Table 4). The PCR sequencing temperature outline maintained in a 1st cycle at 96oC for 2 minutes followed by 30 cycles at 96oC for 30 sec, 50oC for 40 sec and 60oC for 4 minutes for all the primers (Table 5).
Table 4: PCR for DNA sequence amplification conditions
|
Constituents |
Quantity |
|
PCR Product (ExoSAP treated) |
10-20ng |
|
Primer |
3.2pM (either Forward or Reverse) |
|
Sequencing Mix |
0.28µl |
|
5x Reaction buffer |
1.86µl |
|
Sterile distilled water |
make up to 10µl |
Table-5: PCR Temperature profile for DNA sequence
|
Profile |
Specification |
|
Initial denaturation |
96°C for two minutes |
|
Denaturation |
96°C for two minutes |
|
Annealing |
96°C for 30 seconds |
|
Extension |
50°C for 40 seconds |
|
Final extension |
60°C for 4 minutes |
|
Number of cycles |
30cycles |
The sequences were analyzed by the BLAST program in the NCBI (National Center for Biotechnology Information) database to confirm the PCR target. The electropherograms of each sequence were tested for sequence correctness using a Sequence Scanner version 1.0 and Bioedit version 7.1. All sequences were aligned automatically using Clustal W version 2.0.
The rbcL sequences of Gracilaria fergusonii sample were determined by direct sequencing. Phylogenetic relationships within and among Gracilaria specimens were analyzed based on rbcL sequence data using the Neighbor-joining method and UPGMA method. Twenty-nine isolates of Gracilaria from the Genbank database were used to construct the phylogenetic tree (Table 6).
Table 6: List of Gracilaria species and sequences of rbcL used for constructed phylogenetic analysis.
|
Gracilaria Species |
Reference |
|
Gracilaria tenuistipitata |
AY049324.1 |
|
Gracilaria vermiculophylla |
JQ768774.1 |
|
Gracilaria cuneata |
EU380717.1 |
|
Gracilaria domingensis |
AY049371.1 |
|
Gracilaria capensis |
AY049378.1 |
|
Gracilaria beckeri |
AY049377.1 |
|
Gracilaria stipitata |
HQ896851.1 |
|
Gracilaria spinulosa |
AY049395.1 |
|
Gracilaria viellardii |
AY049394.1. |
|
Gracilaria taiwanensis |
HQ896848.1 |
|
Gracilaria vieillardii |
HQ896855.1 |
|
Gracilaria flabelliforme |
AY049343.1 |
|
Gracilaria ornate |
AY049318.1 |
|
Gracilaria fergusonii |
KY115201.1 |
|
Gracilaria mammillaris |
AY049323.1 |
|
Gracilaria venezuelensis |
AF539603.1 |
|
Gracilaria babae |
KF831125.1 |
|
Gracilaria arcuata |
AY049383.1 |
|
Gracilaria dura |
GQ229499.1 |
|
Gracilaria gracilis |
AY049400.1 |
|
Gracilaria gracilis |
GQ229500.1 |
|
Gracilaria pacifica |
AY049397.1 |
|
Gracilaria crassissima |
AY049351.1 |
|
Gracilaria truncate |
HQ896852.1 |
|
Gracilaria urvilleii |
AY049402.1 |
|
Gracilaria cylindrica |
KY115202.1 |
|
Gracilaria chilensis |
KP857578.1 |
|
Gracilaria salicornia |
EU937772.1 |
|
Gracilaria corticata |
EU937767.1 |
In the present study, the genomic DNA from Gracilaria fergusonii J.Ag. was isolated and amplified using rbcL primers and the amplified DNA was sequenced. The partial rbcL gene sequence of Gracilaria fergusonii J.Ag. was deposited to GenBank of NCBI and was assigned with the accession number of KY115201.1.
In the present study, the chromatogram of rbcL barcoding gene sequences of the selected Grailaria fergusonii (Figure 1) was generated.
Figure 1: DNA sequence chromatogram of Gracilaria fergusonii (rbcL)
The nucleotide composition of rbcL gene sequences of the selected Gracilaria fergusonii species was analyzed using the BioEdit software version 5.0.6 and the results were illustrated in Figure 2. 927 nucleotides was displayed in Gracilaria fergusonii collected from Hare Island in Thoothukudi district. The total percentages of adenine, guanine, thymine, and cytosine content were 26.43%, 19.85%, 27.83%, and 25.89% in Gracilaria fergusonii (Table 7 and Figure 3).

Figure 2: Nucleotide sequence of Gracilaria fergusonii
Table 7: Nucleotide compositions of Gracilaria fergusonii.
|
S.No. |
Nucleotide |
Total Number |
Percentage |
|
1. |
A |
287 |
26.43 |
|
2. |
C |
230 |
25.89 |
|
3. |
G |
272 |
19.85 |
|
4. |
T |
286 |
27.83 |
|
Total: 927 |
|||
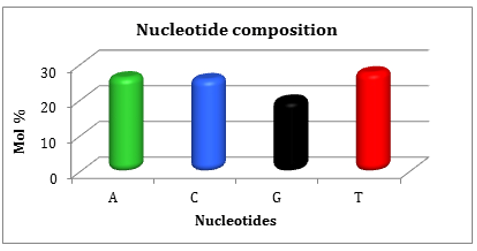
Figure 3: Nucleotid compositions of Gracilaria fergusonii express percentage wise
The phylogenetic analysis of the selected Gracilaria species was performed using different types of Bootstrap analysis of rbcL gene sequences viz., Neighbor-joining method, and UPGMA method. The various rbcL gene sequences retrieved from NCBI and the percentage of similarity and evolutionary distance was calculated using the branch length of the cladogram presented (Figure 4).

Figure 4: Multiple Sequence Alignment Dendrogram of overall Gracilaria species
This study deals with resolving the taxonomical problem of red algae utilizing rbcL sequence; it is a reliable tool when species identification is required quickly. The ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) large subunit gene (rbcL) was chosen for the DNA barcoding as the target of this assay for several reasons. rbcL is the most profuse enzyme on the globe and has fascinated much phylogenetic consideration, rbcL catalyzes the assimilation of carbon dioxide to organic carbon through the Calvin-Benson cycle. DNA barcoding and molecular taxonomy of Gracilaria fergusonii J.Ag. using rbcL primers with a sequence of 927 bp were successfully carried out. The Gracilaria fergusonii J.Ag. have a similarity of 99% genetically with other Gracilaria fergusonii J.Ag. in the world.
The authors are thankful to GeneBank, NCBI for accepting the partial sequence of rbcL gene of Gracilaria cylindrica Boergesen and providing the accession number: KY115201.1.
REFERENCES
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.